About the CoMSES Model Library more info
Our mission is to help computational modelers at all levels engage in the establishment and adoption of community standards and good practices for developing and sharing computational models. Model authors can freely publish their model source code in the Computational Model Library alongside narrative documentation, open science metadata, and other emerging open science norms that facilitate software citation, reproducibility, interoperability, and reuse. Model authors can also request peer review of their computational models to receive a DOI.
All users of models published in the library must cite model authors when they use and benefit from their code.
Please check out our model publishing tutorial and contact us if you have any questions or concerns about publishing your model(s) in the Computational Model Library.
We also maintain a curated database of over 7500 publications of agent-based and individual based models with additional detailed metadata on availability of code and bibliometric information on the landscape of ABM/IBM publications that we welcome you to explore.
Displaying 10 of 875 results for "P Van Geert" clear search
Population Dynamics of Emerald Ash Borer
mpeters | Published Monday, December 13, 2010 | Last modified Saturday, April 27, 2013This model was developed as part of a class project, and explores the population dynamics and spread of an invasive insect, Emerald Ash Borer, in a county.
Investigating dynamics of covid-19 spread and containment with agent-based modeling
Amirarsalan Rajabi | Published Thursday, January 21, 2021This repository includes an epidemic agent-based model that simulates the spread of Covid-19 epidemic. Normal.nlogo is the main file, while Exploring-zoning.nlogo and Exploring-Testing-With-Tracking.nlogo are modefied models to test the two strategies and run experiments.
Pastoralscape
Matthew Sottile | Published Tuesday, October 12, 2021Pastoralscape is a model of human agents, lifestock health and contageous disease for studying the impact of human decision making in pastoral communities within East Africa on livestock populations. It implements an event-driven agent based model in Python 3.
Charcoal Record Simulation Model (CharRec)
Grant Snitker | Published Monday, November 16, 2015 | Last modified Thursday, September 30, 2021This model (CharRec) creates simulated charcoal records, based on differing natural and anthropogenic patterns of ignitions, charcoal dispersion, and deposition.
Between Pleasure and Contentment: Evolutionary Dynamics of Some Possible Parameters of Happiness
Yue Gao Shimon Edelman | Published Saturday, March 12, 2016 | Last modified Wednesday, March 16, 2016We build a computational model to investigate, in an evolutionary setting, a series of questions pertaining to happiness.
Machine Learning simulates Agent-based Model
B Furtado | Published Wednesday, March 07, 2018This is an initial exploratory exercise done for the class @ http://thiagomarzagao.com/teaching/ipea/ Text available here: https://arxiv.org/abs/1712.04429v1
The program:
Reads output from an ABM model and its parameters’ configuration
Creates a socioeconomic optimal output based on two ABM results of the modelers choice
Organizes the data as X and Y matrices
Trains some Machine Learning algorithms
…
St Anthony flu
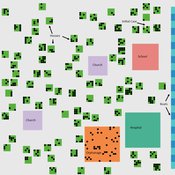
Lisa Sattenspiel | Published Monday, April 15, 2019The St Anthony flu model is an epidemiological model designed to test hypotheses related to the spread of the 1918 influenza pandemic among residents of a small fishing community in Newfoundland and Labrador. The 1921 census data from Newfoundland and Labrador are used to ensure a realistic model population; the community of St. Anthony, NL, located on the tip of the Northern Peninsula of the island of Newfoundland is the specific population modeled. Model agents are placed on a map-like grid that consists of houses, two churches, a school, an orphanage, a hospital, and several boats. They engage in daily activities that reflect known ethnographic patterns of behavior in St. Anthony and other similar communities. A pathogen is introduced into the community and then it spreads throughout the population as a consequence of individual agent movements and interactions.
A Simulation of Arab Spring Protests Informed by Qualitative Evidence
Bruce Edmonds Stephanie Dornschneider | Published Monday, April 29, 2019 | Last modified Friday, May 24, 2019The purpose of the simulation was to explore and better understand the process of bridging between an analysis of qualitative data and the specification of a simulation. This may be developed for more serious processes later but at the moment it is merely an illustration.
This exercise was done by Stephanie Dornschneider (School of Politics and International Relations, University College Dublin) and Bruce Edmonds to inform the discussion at the Lorentz workshop on “Integrating Qualitative and Quantitative Data using Social Simulation” at Leiden in April 2019. The qualitative data was collected and analysed by SD. The model specification was developed as the result of discussion by BE & SD. The model was programmed by BE. This is described in a paper submitted to Social Simulation 2019 and (to some extent) in the slides presented at the workshop.
Classical Swine Fever in wild boars
Cédric Scherer Martin Lange Volker Grimm Hans-Hermann Thulke Stephanie Kramer-Schadt | Published Friday, September 06, 2019The model is a combination of a spatially explicit, stochastic, agent-based model for wild boars (Sus scrofa L.) and an epidemiological model for the Classical Swine Fever (CSF) virus infecting the wild boars.
The original model (Kramer-Schadt et al. 2009) was used to assess intrinsic (system immanent host-pathogen interaction and host life-history) and extrinsic (spatial extent and density) factors contributing to the long-term persistence of the disease and has further been used to assess the effects of intrinsic dynamics (Lange et al. 2012a) and indirect transmission (Lange et al. 2016) on the disease course. In an applied context, the model was used to test the efficiency of spatiotemporal vaccination regimes (Lange et al. 2012b) as well as the risk of disease spread in the country of Denmark (Alban et al. 2005).
References: See ODD model description.
Agent-based model of WiFi tracking system in urban environment
Christopher Thron Khoi Tran | Published Friday, April 21, 2017This code simulates the WiFi user tracking system described in: Thron et al., “Design and Simulation of Sensor Networks for Tracking Wifi Users in Outdoor Urban Environments”. Testbenches used to create the figures in the paper are included.
Displaying 10 of 875 results for "P Van Geert" clear search