MHMSLeptoDy (Multi-host, multi-serovar Leptospira Dynamics Model) 1.1.2
Leptospirosis is a neglected, bacterial zoonosis with worldwide distribution, primarily a disease of poverty. More than 200 pathogenic serovars of Leptospira bacteria exist, and a variety of species may act as reservoirs for these serovars. Human infection is the result of direct or indirect contact with Leptospira bacteria in the urine of infected animal hosts, primarily livestock, dogs, and rodents. There is increasing evidence that dogs and dog-adapted serovar Canicola play an important role in the burden of leptospirosis in humans in marginalized urban communities. What is needed is a more thorough understanding of the transmission dynamics of Leptospira in these marginalized urban communities, specifically the relative importance of dogs and rodents in the transmission of Leptospira to humans. This understanding will be vital for identifying meaningful intervention strategies.
One of the main objectives of MHMSLeptoDy is to elucidate transmission dynamics of host-adapted Leptospira strains in multi-host system. The model can also be used to evaluate alternate interventions aimed at reducing human infection risk in small-scale communities like urban slums.
Release Notes
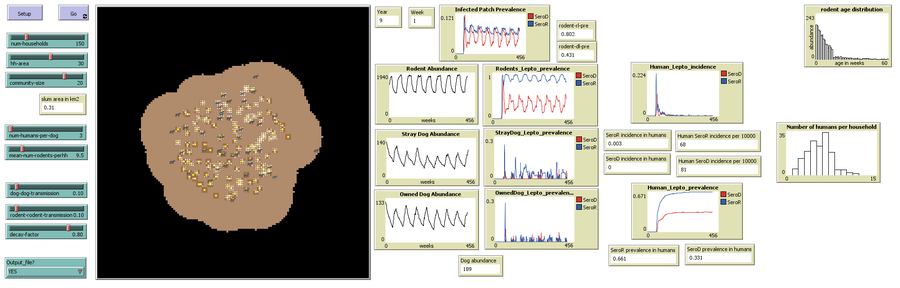
The output file ( now titled ‘HostLeptoIncidence.csv’) records four additional outputs for every year of model run: i) annual number of cases due to dog serotype in dogs, ii) annual number of cases due to rodent serotype in dogs, iii) annual number of cases due to rodent serotype in rodents, and iv) annual number of cases due to dog serotype in rodents.
1) The user specifies the number of households desired (slider ‘num-households’), household area (slider ‘hh-area’: neighboring patches belonging to/influenced by a household) and the community-area (slider ‘community-size’: area of influence of all the households).
2) The user sets the initial dog and rodent population using two sliders: ‘num-humans-per-dog‘ and ‘mean-num-rodents-perhh’.
3) The probabilities of direct transmission are set using sliders ‘rodent-rodent-transmission‘ and ‘dog-dog-transmission’. The proportion of leptospires surviving each day is set using slider ‘decay-factor’.
4) If model outputs are to be stored as a .csv file, select ‘YES’ using the dropdown menu for the chooser ‘Output-file?’